 Infographic by Linlin Zhao, University of California Riverside
Infographic by Linlin Zhao, University of California Riverside
 Infographic by Linlin Zhao, University of California Riverside
Infographic by Linlin Zhao, University of California Riverside
Studies of ancient DNA have tended to focus on frozen land in the northern hemisphere, where woolly mammoths and bison roamed. Meanwhile, Antarctica has received relatively little attention. We set out to change that.
The most suitable sediments are exposed near the coast of the icy continent, where penguins like to breed. Their poo is a rich source of DNA, providing information about the health of the population as well as what penguins have been eating.
Our new research opens a window on the past of Adélie penguins in Antarctica, going back 6,000 years. It also offers a surprise glimpse into the shrinking world of southern elephant seals over the past 1,000 years.
Understanding how these species coped with climate change in the past can help us prepare for the future. Wildlife in Antarctica faces multiple emerging threats and will likely need support to cope with the many challenges ahead.
Adélie penguins are particularly sensitive to changes in their environment. This makes them what we call a “sentinel species”, providing an early warning of imbalance or dysfunction in the coastal ecosystem. Their poo also provides a record of how they responded to changes in the past.
In our new research, we excavated pits up to 80cm deep at ten Adélie penguin colonies along the 700km Ross Sea coastline. We then collected 156 sediment samples from different depths in each excavation.
Six of these colonies were still active, meaning birds return annually to breed. The other four had been abandoned at various times over the past 6,000 years.
From these sediments we generated 94 billion DNA sequences, which provided us with an unparalleled window into the past lives of Adélie penguins and their ecosystem.
We detected the DNA of several animal species besides Adélie penguins. These animals included two other birds, three seals and two soil invertebrates.
Not all of this DNA came from penguin poo. Our samples also contained DNA from feathers, hairs or skin cells of other species in the environment at the time.
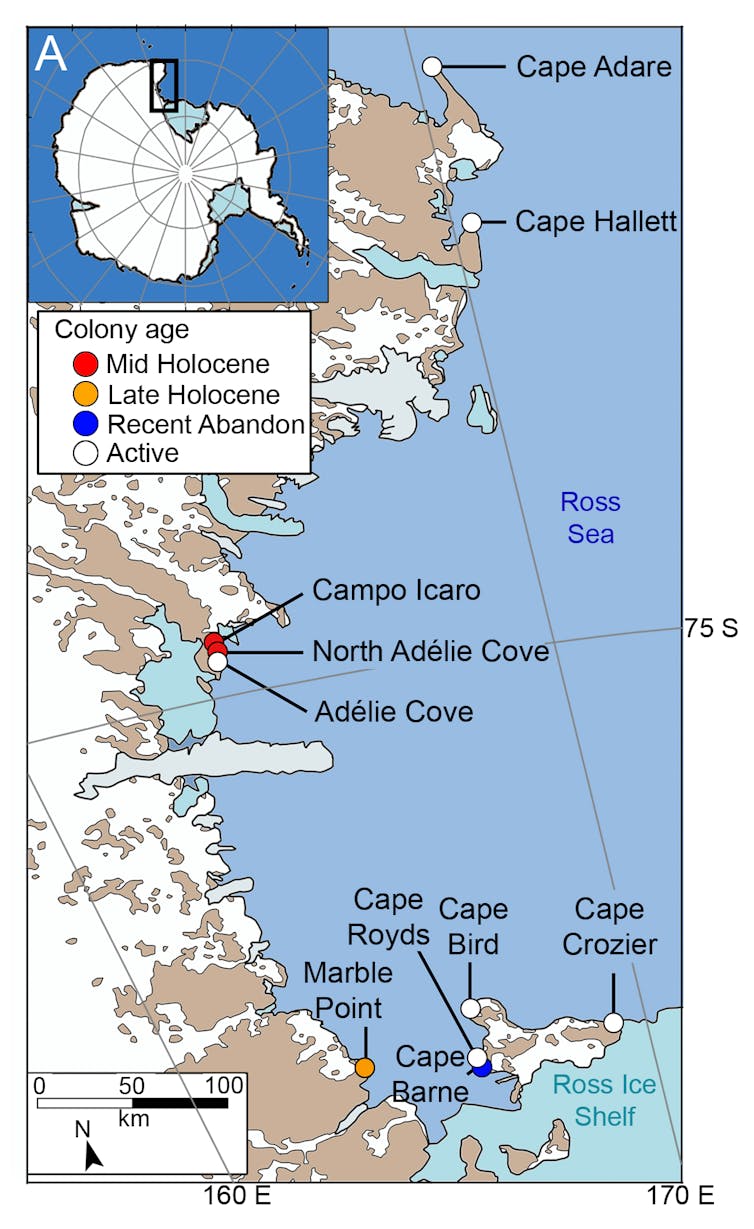 Sediment samples were taken from ten penguin colonies of various ages, six active (white dot) and four abandoned (coloured dot), on the coast of Ross Sea in Antarctica. Wood, J., et al (2025) Nature Communications, CC BY-NC-ND
Sediment samples were taken from ten penguin colonies of various ages, six active (white dot) and four abandoned (coloured dot), on the coast of Ross Sea in Antarctica. Wood, J., et al (2025) Nature Communications, CC BY-NC-NDWhen we took a closer look at the DNA from penguins of the present day, we found more genetic diversity in samples from larger colonies.
Recognising this relationship between genetic diversity and colony size enabled us to estimate the size of former colonies. We could also reconstruct population trends through time.
For example, in samples from active colonies, we found penguin genetic diversity increased as we sampled closer and closer to the surface. This may reflect population growth over the past century.
The DNA also revealed changes in penguin diets over time. Over the past 4,000 years, the penguins in the southern Ross Sea switched from mainly eating one type of fish – the bald notothen – to another, Antarctic silverfish.
The bald notothen lives beneath the sea ice, so this prey-switching was likely driven by a change in sea ice extent compared with the past.
We made an unexpected discovery at Cape Hallett, in the northern Ross Sea. This is the site of an active penguin colony.
Samples of sediment from close to the surface contained lots of penguin DNA and eggshell. But samples from further down, where penguin DNA and eggshell were scarce, contained DNA from southern elephant seals.
Today, elephant seals are uncommon visitors to the Antarctic continent, and breed on subantarctic islands including Macquarie, Campbell and Antipodes Islands. Yet, bones of elephant seal pups found along the Ross Sea coast indicate the species used to breed in the area.
Carbon dating of these bones indicate elephant seal colonies began disappearing from the southern Ross Sea around 1,000 years ago. Over the following 200 years, colonies in the northern Ross Sea began vanishing too.
As the climate cooled and the extent of sea ice increased, elephant seals could no longer access suitable breeding sites. These sites were then taken over by Adélie penguins who expanded into areas once occupied by seals.
Our DNA evidence suggests Cape Hallett was one of the last strongholds of southern elephant seals on the icy continent. But we may yet again see elephant seals breeding on the Antarctic mainland as the world warms and sea ice melts.
Our study spans the past 6,000 years, but our research suggests it would be possible to go even further back.
The DNA fragments we found were very well preserved, showing little of the damage expected in warmer climates.
So it should be possible to obtain much older DNA from sediments on land in Antarctica – maybe even 1 million-year-old DNA, as recently reported from Antarctic sediments beneath the ocean floor.
In December 2017, 2.09 million square kilometres of the Ross Sea and adjoining Southern Ocean became the world’s largest marine protected area. Establishing the protection was a major achievement, yet it was only afforded for 35 years.
After 2052, continuation of the region’s protected status will require international agreement. Knowledge of the vulnerability of local species and their risk in the face of change will play a key role in informing the decision. Our research provides a case study for how ancient environmental DNA can contribute towards this understanding.![]()
Jamie Wood, Senior Lecturer in Ecology and Evolution, University of Adelaide and Theresa Cole, Postdoctoral technician in environmental DNA, University of Adelaide
This article is republished from The Conversation under a Creative Commons license. Read the original article.


 Scientists in Kashmir have cloned the first Pashmina goat using advanced reproductive techniques, officials at the Shere- Kashmir University of Agricultural Sciences &Technology (SKUAST) said on Thursday. The March 9 birth of female kid Noori could spark breeding programmes across the region and mass production of the highpriced wool, lead project scientist Dr Riaz Ahmad Shah said. Shah and six other scientists took two years to clone Noori, using the relatively new “ handmade” cloning technique involving only a microscope and a steady hand. “ We’ve standardised the procedure. Now it will take us half a year to produce another,” said Dr Maajid Hassan, another veterinarian who worked on the project, which was partly funded by the World Bank. The team has already started work on more clones among the university’s herd of goats. “ This is the cheapest, easier and less time- consuming” method of cloning, compared with conventional methods that use high- tech machinery and sometimes chemicals, Shah said. Pashmina, a kind of fine wool is obtained from the fleece of the goat Capra hircus . They are found in parts of the Himalayas, the Tibetan plateau and upper reaches of Ladakh. The wool is spun through a tedious manual processto produce the finest quality of Pashmina. Source: Ananta-Tech
Scientists in Kashmir have cloned the first Pashmina goat using advanced reproductive techniques, officials at the Shere- Kashmir University of Agricultural Sciences &Technology (SKUAST) said on Thursday. The March 9 birth of female kid Noori could spark breeding programmes across the region and mass production of the highpriced wool, lead project scientist Dr Riaz Ahmad Shah said. Shah and six other scientists took two years to clone Noori, using the relatively new “ handmade” cloning technique involving only a microscope and a steady hand. “ We’ve standardised the procedure. Now it will take us half a year to produce another,” said Dr Maajid Hassan, another veterinarian who worked on the project, which was partly funded by the World Bank. The team has already started work on more clones among the university’s herd of goats. “ This is the cheapest, easier and less time- consuming” method of cloning, compared with conventional methods that use high- tech machinery and sometimes chemicals, Shah said. Pashmina, a kind of fine wool is obtained from the fleece of the goat Capra hircus . They are found in parts of the Himalayas, the Tibetan plateau and upper reaches of Ladakh. The wool is spun through a tedious manual processto produce the finest quality of Pashmina. Source: Ananta-Tech Everybody is talking about internet addiction – many people spend hours online and immediately start feeling bad if they are unable to do so. Medically, this phenomenon has not been as clearly described as nicotine or alcohol dependency. But a German study in the Journal of Addiction Medicine suggests that there are molecular-genetic connections in internet addiction, too. "It was shown that Internet addiction is not a figment of our imagination," says the lead author, Dr. Christian Montag of the University of Bonn. "Researchers and therapists are increasingly closing in on it." He found that some people’s thoughts revolve around the internet during the day and that they feel their wellbeing is severely impacted if they have to go without it. The problem users seem to have a genetic variation that also plays a major role in nicotine addiction. "It seems that this connection is not only essential for nicotine addiction, but also for internet addiction," reports the Bonn psychologist. “The current data already shows that there are clear indications for genetic causes of Internet addiction." The actual mutation is on the CHRNA4 gene that changes the genetic make¬up for the Alpha 4 subunit on the nicotinic acetylcholine receptor. "Within the group of subjects exhibiting problematic Internet behavior this variant occurs more frequently – in particular, in women," says Dr. Montag. ~ Universität Bonn press release, Aug 29 Source BioEdge, Image
Everybody is talking about internet addiction – many people spend hours online and immediately start feeling bad if they are unable to do so. Medically, this phenomenon has not been as clearly described as nicotine or alcohol dependency. But a German study in the Journal of Addiction Medicine suggests that there are molecular-genetic connections in internet addiction, too. "It was shown that Internet addiction is not a figment of our imagination," says the lead author, Dr. Christian Montag of the University of Bonn. "Researchers and therapists are increasingly closing in on it." He found that some people’s thoughts revolve around the internet during the day and that they feel their wellbeing is severely impacted if they have to go without it. The problem users seem to have a genetic variation that also plays a major role in nicotine addiction. "It seems that this connection is not only essential for nicotine addiction, but also for internet addiction," reports the Bonn psychologist. “The current data already shows that there are clear indications for genetic causes of Internet addiction." The actual mutation is on the CHRNA4 gene that changes the genetic make¬up for the Alpha 4 subunit on the nicotinic acetylcholine receptor. "Within the group of subjects exhibiting problematic Internet behavior this variant occurs more frequently – in particular, in women," says Dr. Montag. ~ Universität Bonn press release, Aug 29 Source BioEdge, Image

 Fish provide clues for how environmental factors can lead to vision changes. Photo of scorpionfish by Andrew David, NOAA's Fisheries Collection.
Fish provide clues for how environmental factors can lead to vision changes. Photo of scorpionfish by Andrew David, NOAA's Fisheries Collection. Unlike many other animals, most primates, including humans, have both a red and a green pigment, enabling them to distinguish red from green and vice-versa. Photo by Richard Ruggiero, U.S. Fish and Wildlife Service.
Unlike many other animals, most primates, including humans, have both a red and a green pigment, enabling them to distinguish red from green and vice-versa. Photo by Richard Ruggiero, U.S. Fish and Wildlife Service.